Methylomics offers biomarker discovery as a service
Target cell enrichment for biomarker discovery
For biomarker discovery, enriched target cell populations are often used. For formalin-fixed and paraffin-embedded tissue samples the enrichment is done by laser capture micro-dissection (LCM). Sections of FFPE tissue are prepared on coated glass slides, dewaxed, dehydrated, and stained with Hematoxylin. Slides are scanned using an imager and the regions of interest are annotated by a pathologist. The regions are then laser-cut using a LCM system, followed by a genomic DNA extraction. DNA concentrations are measured and the samples are prepared for MeD-seq analysis.
Figure 1. isolation of target cells using Laser Capture Microdisseciton (LCM).
Differently Methylated Regions
With the MeD-seq data we construct genome-wide DNA methylation profiles of normal and diseased tissue types (Figure 2).
Comparison of cancer and control methylation profiles by an in house developed bioinformatics pipeline facilitates the identification of specific or general Differentially Methylated Regions (DMR) in a genome wide fashion (Figure 3).
Our pipeline is capable of detecting the following types of DMR’s
- General, for example regions methylated in all cancer types of the female genital zone
- Regional, for example methylated in 2 or more cancer types (Cervix and Vulva carcinoma)
- Specific, for example methylated in one specific cancer type (Cx-AdC or Vul-SCC HPV negative)
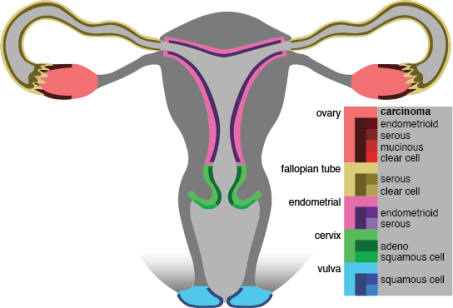
Figure 2. female urogenital zone, methylations profiles are generated for both control and cancer tissue samples.

Figure 3. illustrative representation of IGV captions of the 5 DMR types: Two general markers where all cancers have methylation reads and all controls do not have methylation reads. One regional marker with high methylation for cervix and vulva cancers, low methylation for cervical and vulva controls, all other controls and all other cancers. Two specific regions; one region with only methylation for cervical adenocarcinoma samples and one region with only methylation for vulva squamous cell carcinoma HPV negative samples.
Assay design and primer and probe selection
From the DMR regions Methylomics develops specific, regional and general biomarkers. However, not all DMR’s are suitable to create powerful cancer detection biomarkers. To determine whether a DMR is discriminating enough and suitable to create a powerful marker, quality scores, predictive scores and visualizations are applied to select candidate biomarkers for assay development.
Visualization of these selected DMR’s in grouped DNA methylation profiles through an Integrative Genomics Viewer (IGV) facilitates the identification of promising target sequences for the development of quantitative Methylation-Specific PCR (qMSP) assays (Figure 3).
Methylomics develops multiplex assays detecting the methylation status of multiple regions at once. To assess the potential of the newly discovered markers the developed assays are tested on bisulfite-converted DNA or methylation-sensitive restriction enzyme (MSRE) cutted DNA of relevant samples.
